Background
What is computational biology?
Computational biology (“comp bio”) involves using advanced computational techniques to answer biological questions. Have you heard of experiments performed in vivo or in vitro? More and more, scientists use simulations and big-data analyses to perform experiments in silico.
What does computational biology study?
Comp bio sheds light on biology’s “central dogma”. To simplify:
- DNA is like a book, and each chapter (“gene”) describes how to make one of our body’s many proteins.
- RNA-polymerase (a protein) copies a given chapter, producing a shorter strand of messenger RNA. RNA strands are chemically similar to DNA, but each strand contains the instructions for a single protein.
- A large molecule called the ribosome reads the copy and makes the corresponding protein.
- The protein then performs its cellular function, speeding up a chemical reaction, helping to relay a message from outside the cell, etc.
Sometimes the central dogma goes wrong. Mistakes in the DNA code get passed to the messenger RNA and ultimately to the protein, causing genetic diseases. Computational biology can also study bacteria, fungi, and viruses, which all use DNA, RNA, and proteins too.
Are there different kinds of comp bio?
Computational biology includes two broad subfields: bioinformatics and computational structural biology. Bioinformatics focuses mostly on the DNA and RNA code. Computational structural biology considers the 3D structures and motions of proteins and interacting small molecules.
What kind does the Durrant lab study?
Our research touches on both subfields, but mostly we’re computational structural biologists. Our projects center on small-molecule ligand identification and molecular visualization.
Small-Molecule Ligand Identification
What is a small-molecule ligand?
Ligands are chemicals that bind to grooves and pockets in viral, bacterial, parasitic, and cancer-related proteins. Ligand binding changes protein function, thus interfering with associated disease/biological processes.
Why do you care about ligands?
Ligands serve both medical and basic-science purposes. Some can be developed into new drugs to treat human disease. Others are used as chemical probes to study proteins’ biological roles. Computer-aided drug design (CADD) accelerates ligand identification by predicting protein binding in a computer, allowing scientists to test fewer molecules before finding one that’s effective.
CADD will further “personalized medicine,” which aims to provide patient-specific treatments based on unique personal genetics. CADD-accelerated drug discovery is critical given the great variety of customized medicines required.
What techniques do you develop and use?
Computer docking/scoring is a common CADD method used to identify new protein-binding molecules. It has three steps:
- Docking: Predict the correct position/orientation (or “pose”) that a ligand assumes when binding to a protein pocket.
- Scoring: From the ligand’s 3D geometry, calculate a score that indicates how strongly it binds to the protein. Effective drugs/probes usually bind strongly.
- Experimental validation: If a given ligand has a good score, test it in an actual experiment to verify binding.
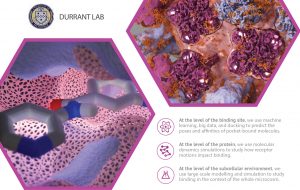
Researchers use docking/scoring extensively and successfully, but it is far from 100% accurate. Improving accuracy will lead to higher experimental “hit rates,” further accelerating early-stage discovery. The Durrant lab uses machine learning and big-data databases to improve both the docking and scoring aspects of this process.
We also use molecular dynamics simulations to study protein motions. Proteins are in constant motion, so their surface pockets constantly change shape. Any one of these many pocket “conformations” might be compatible with drug binding. CADD techniques that focus on a single pocket conformation–ignoring the many others–might miss some effective ligands.
We develop and apply methods to analyze these simulations. Because nearby proteins and lipid bilayers also influence protein motions, we also perform larger-scale simulations of entire subcellular environments.
Molecular Visualization
What is molecular visualization?
Proteins and ligands are invisible to the eye, so visualizing comp-bio results requires modeling, scientific illustration, and 3D artistry. Molecular visualizations provide scientific insights and can serve as powerful educational tools.
How do you make protein pictures?
Many programs contribute to a high-quality molecular visualization. Visual molecular dynamics (VMD) displays docked molecules and simulations on a computer screen. We import these on-screen models into Blender, a 3D computer-graphics program used in the film and video-game industries. Blender provides careful control over lighting and materials, allowing for photo-realistic images. Beyond creating these visualizations, Durrant-lab students also develop techniques to improve molecular graphics.
What if I want to walk around a protein?
We’re developing virtual-reality (VR) software so we can walk around–and through–our favorite proteins. ProteinVR will be a useful educational and collaborative tool. Students involved in this project use many of the techniques common to the video-game industry.