Many of our research tools have been implemented as easy-to-use browser apps that are well suited for even large classroom settings. Please let us know if you use any of our tools in your courses!

BINANA
BINANA (BINding ANAlyzer) analyzes the geometries of predicted ligand poses to identify molecular interactions that contribute to binding. It is useful because accurately characterizing these interactions allows medicinal chemists to assess whether a predicted ligand merits further study. We have also created a BINANA web-browser application so students and chemical-biology researchers can quickly visualize receptor/ligand complexes and their unique binding interactions.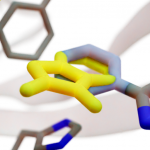
DeepFrag
DeepFrag is a deep convolutional neural network that guides ligand optimization by extending a ligand with a molecular fragment, such that the resulting extension is also highly complementary to the receptor. The DeepFrag web application is also a useful tool for teaching students about medicinal chemistry and lead optimization.
FPocketWeb
FpocketWeb is a browser app for identifying pockets on protein surfaces where small-molecule ligands (e.g., drugs) might bind. It runs the fpocket executable entirely in a web browser. The pocket-finding calculations occur on the user’s computer rather than a remote server.
PCAViz
PCAViz is an open-source Python/JavaScript toolkit for sharing and visualizing MD trajectories via a web browser. It is also a helpful tool for class websites and presentations. To encourage use, an easy-to-install PCAViz-powered WordPress plugin enables ‘plug-and-play’ trajectory visualization.
Prot2Prot
Prot2Prot is a deep-learning model trained to imitate a Blender-rendered image given a much simpler representation (‘sketch’) of a protein surface. Prot2Prot outputs an image that is often indistinguishable from a Blender/BlendMol-based visualization in a fraction of the time, allowing image ‘rendering’ even in a web browser.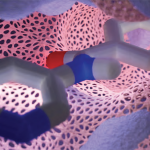
ProteinVR
ProteinVR is a web-based application that allows users to view protein/ligand structures in virtual reality (VR) from their mobile, desktop, or VR-headset-based web browsers. Molecular structures are displayed within 3D environments that give useful biological context and allow users to situate themselves in 3D space. We expect the ProteinVR web application to be useful in both research and classroom settings.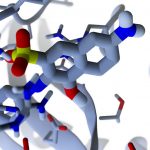
Webina
Webina is a JavaScript/WebAssembly library that runs AutoDock Vina entirely in a web browser. The docking calculations take place on the user’s own computer rather than a remote server. To encourage use, we have incorporated the Webina library into our own Webina web application. The app includes a convenient interface so users can easily setup their docking runs and analyze the results. Aside from its research applications, Webina is an excellent tool for teaching students about computer-aided drug discovery, protein/ligand interactions, and computational structural biology.

